Varroa mite genomes resource and preprint
Over the past decades, the ectoparasitic Varroa mite has been the key driver of honey bee population declines worldwide. Surprisingly, no modern genome reference existed for the global beekeeping pest V. destructor, nor any other Varroidae. We developed new reference genomes for V. destructor and its sister species V. jacobsoni which also jumped last decade, from the eastern honey bee Apis cerana to the western honey bee A. mellifera. Using a combination of HiSeq, PacBio, and Hi-C sequencing, we obtained a genome at a chromosomal-level for V. destructor with the best N50 scaffold for any arachnid to date.
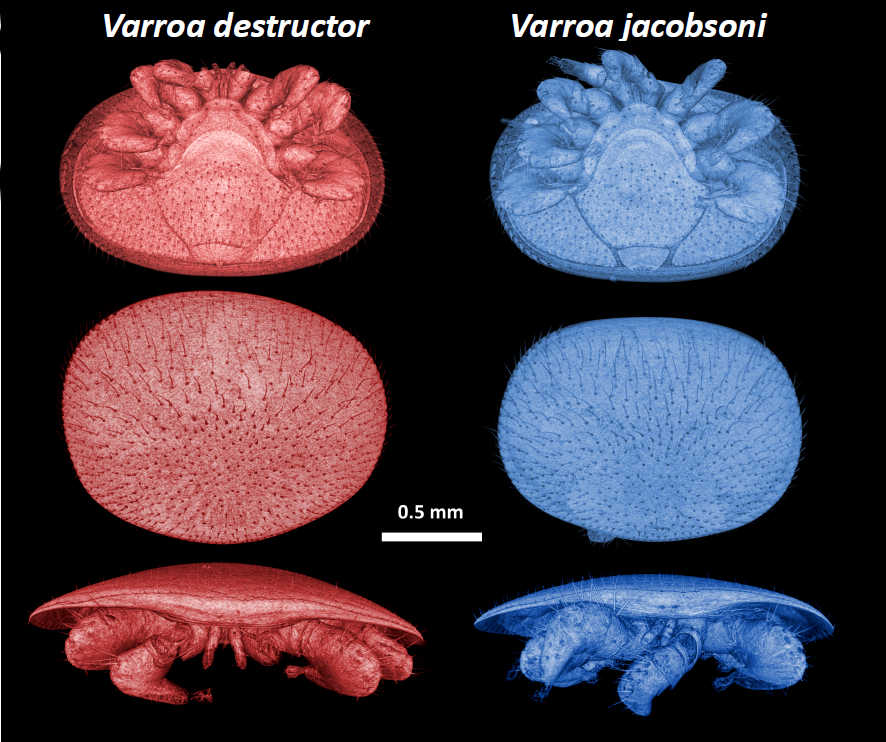
Both have been on NCBI for a while now, but we finally posted our comparative genomic analysis of these two Varroa mites preprint on bioarxiv. We found out that although both Varroa species have the same host range and specificity, they show striking differences in genes under selection and divergent strategies of adaptation. If you want to learn more about these nasty little beasts, you can read our work here: Genomic analyses of sibling honey bee ectoparasitic mite species show divergent strategies of adaptation, until hopefully a more formal publication soon.
We hope these resource will help better understanding the success of these mites and open new paths for species-specific pest control.
